(click on images!)
Simultaneous multi-scale electrophysiological measurement and optical manipulation of in vivo cortical networks
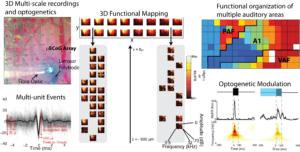
We pioneered a 3D electrophysiological recording system that combines micro-eletrocorticography (uECoG) to record neural activity from the cortical surface over extended areas with mesoscale spatial resolution, with laminar polytrodes to densely record neural activity across cortical layers with microscale spatial resolution. Combining these high-temporal resolution, multiscale electrophysiological recordings with optical manipulations of neural activity further allows causal inference into the role of specific neural populations in local and distributed cortical computations. Examination of recorded signals >1kHz reveals neural events with timing and amplitude characteristics indicative of multi-unit action potentials evoked by sounds. Direct recordings of action potentials with laminar polytrodes inserted through perforations in the uECoG array suggest that functional tuning derived from uECoG (70-170Hz) reflects a spatial average of multiunit spiking activity immediately beneath the uECoG contacts. Focusing on the activity in the high-gamma range, we demonstrated that uECoG recorded field potentials have sufficient spatial resolution and selectivity to derive functional organization of rat auditory cortex across multiple cortical areas simultaneously(tonotopy), and thus provide a method for rapid, non-destructive mapping of cortical function. Preliminary results demonstrate the ability of uECoG to record neural changes in sound-evoked neural activity with high temporal resolution during optical manipulation of specific neuronal populations. Together, these results demonstrate high-temporal resolution, multi-scale electrophysiological measurements with simultaneous optical manipulation of in vivo cortical networks.
Related Publications:
- Strategies for optical control and simultaneous electrical readout of extended cortical circuits. Ledochowitsch, P.*, Yazdan-Shahmorad, A.*, Bouchard, K.E., Diaz-Botia, C., Hanson, T., He, J., Seybold, B., Olivero, E., Blanche, T.J., Schreiner, C.E., Hasenstaub, A., Chang, E.F., Sabes, P.**, Maharbiz, M.M.**; J. Neurosci. Methods, July, 2015.
- Laminar origin of evoked ECoG high-gamma activity. Dougherty, Maximilian E., Nguyen, Anh P. Q., Baratham, Vyassa L., Bouchard, K.E.; 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC) (2019).
- Neural optoelectrodes merging semiconductor scalability with polymeric-like bendability for low damage acute in vivo neuro readout and stimulation. Lanzio, V., Gutierrez, V., Hermiz, J., Bouchard, K.E., Cabrini, S. Journal of Vacuum Science Technology B; 2021
- Columnar localization and laminar origins of Cortical Surface Electrical Potentials. Baratham, V.L., Dougherty, M,E., Hermiz, J., Ledochowitsch, P Maharbiz, M.M., Bouchard, K.E., Journal of Neuroscience, May, 2022.
Statistical-machine learning methods for time-series data
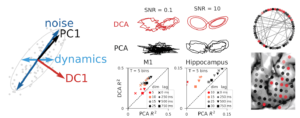
Understanding neural dynamics is key to deciphering the transformations and computations underlying perception, movement, and cognition. However, many of the methods utilized by neuroscientists to understand brain dynamics are not designed to distinguish between noise and true dynamics (far left plot). To this end, we created Dynamic Components Analysis (DCA) to enable unsupervised discovery of dynamically important sub-spaces embedded in high-dimensional, noisy time-series data (e.g., extraction of Lorenz attractor at very low SNR, top-middle). DCA finds subspaces that explicitly maximize the dynamic complexity in the data (define using predictive information). Importantly, DCA outperforms PCA when decoding external variables from population neural recordings (e.g., arm position from motor cortex (M1) and animal location from hippocampus, bottom-middle). This implies that the behaviorally relevant subspaces for neural computations are those with high dynamic complexity, not high variance.
Ultimately, neural dynamics are generated by the interaction of populations of individual neurons. An intuitive and powerful approach to understand these interactions is functional connectomics, which utilizes the formalism of graphs and networks. However, accurate and scalable methods to infer such networks from large scale noise time-series data have previously been lacking. We have shown that our recently introduced Union of Intersections framework (see below) enables extraction of accurate and highly-predictive functional (‘causal’) networks from large-scale time-series data. When applied to neuroscience data (e.g., recordings from human speech motor cortex, far right), the extracted networks are simultaneously more sparse, predictive, and interpretable.
Related Publications:
- Stochastic Collapsed Variational Inference for Structured Gaussian Process Regression Networks. Meng, R., Lee, H., Bouchard, K.E.; Conference of the International Federation of Classification Societies, 2022.
- Achieving Sparsity in Bayesian Vector Autoregressions with Three-Parameter-Beta-Normal Prior. Meng, R., Rangarajan, H., Bouchard, K.E., Seminar on Bayesian Inference in Econometrics and Statistics, 2021
- Sparse and Low-bias Estimation of High Dimensional Vector Autoregressive Models. Ruiz, T., Bhattacharyya, S., Balasubramanian, M., Bouchard, K.E., Learning for Dynamics and Control, 2020.
- Optimizing the Union of Intersections LASSO (UoILASSO) and Vector Autoregressive (UoIVAR) Algorithms for Improved Statistical Inference at Scale. Balasubramanian, M., Ruiz, T., Cook, B., Bhattacharyya, S., Prabhat, Shrivastava, A., Bouchard, K.E.; Submitted to IPDPS’19.
- Unsupervised Discovery of Temporal Structure in Noisy Data with Dynamical Components Analysis. Livezey, J, Clark, D.G., Bouchard, K.E.; Advances in Neural Information Processing Systems 2019.
- Sparse, Predictive, and Interpretable Functional Connectomics with UoI-Lasso. P.S. Sachdeva, S. Bhattacharyya, Bouchard, K.E.; 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC) (2019).
- In vitro Validation of in silico Identified Inhibitory Interactions Liu, H., Bridges., D., Randall, C., Solla, S.A., Wu, B., Hansma, P., Yan, X., Kosik, K.*, Bouchard, K.E.*, Journal of Neuroscience Methods; 2018. *: co-senior authors
- Union of Intersections for Interpretable Data Driven Discovery and Prediction. Bouchard, K.E., et al.; Advances in Neural Information Processing Systems, 2017
Interpretable and predictive statistical-machine learning for science data
 A central goal of neuroscience is to understand how activity in the nervous system is related to features of the external world, or to features of the nervous system itself. A common approach is to model neural responses as a weighted combination of external features, or vice versa. The structure of the model weights can provide insight into neural computations. Often, neural input-output relationships are sparse, with only a few inputs contributing to the output. In part to account for such sparsity, structured regularizers are often incorporated into model fitting optimization. However, by imposing priors, structured regularizers can make it difficult to interpret learned model parameters. The problem of accurately learning model parameters from (noisy) data can be decomposed into two distinct tasks: selection of non-zero features, and estimation of the values of the associated non-zero parameters. We have recently developed the Union of Intersections framework, which performs feature selection through novel mechanism to balances feature selection through compressive intersection operations with feature expansion through union operations. The values of the corresponding selected parameters are then estimated with low-bias and low-variance. In particular, for a variety of model distributions and noise levels, our methods more accurately recover the parameters of sparse models, leading to more parsimonious explanations of outputs, and higher predictive quality on out-of-training data.
A central goal of neuroscience is to understand how activity in the nervous system is related to features of the external world, or to features of the nervous system itself. A common approach is to model neural responses as a weighted combination of external features, or vice versa. The structure of the model weights can provide insight into neural computations. Often, neural input-output relationships are sparse, with only a few inputs contributing to the output. In part to account for such sparsity, structured regularizers are often incorporated into model fitting optimization. However, by imposing priors, structured regularizers can make it difficult to interpret learned model parameters. The problem of accurately learning model parameters from (noisy) data can be decomposed into two distinct tasks: selection of non-zero features, and estimation of the values of the associated non-zero parameters. We have recently developed the Union of Intersections framework, which performs feature selection through novel mechanism to balances feature selection through compressive intersection operations with feature expansion through union operations. The values of the corresponding selected parameters are then estimated with low-bias and low-variance. In particular, for a variety of model distributions and noise levels, our methods more accurately recover the parameters of sparse models, leading to more parsimonious explanations of outputs, and higher predictive quality on out-of-training data.
Related Publications:
- Numerical Characterization of Support Recovery in Sparse Regression with Correlated Design. Kumar, A., Bouchard, K.E.; Communications in Statistics-Simulation and Computation, 2022.
- Improved inference in coupling, encoding, and decoding models and its consequence for neuroscientific interpretation. Sachdeva, P.S., Livezey, J.A., Dougherty, M.E., Gu, B.M., Berke, J.D., Bouchard, K.E., J. Neuroscience Methods, 2021
- Union of Intersections for Interpretable Data Driven Discovery and Prediction. Bouchard, K.E., et al.; Advances in Neural Information Processing Systems, 2017
- UoI-NMFcluster: A Robust Non-negative Matrix Factorization Algorithm for Improved Parts-Based Decompositions from Noisy Data. Ubaru, S., Wu, K.J., Saad, Y., Bouchard, K.E.; International Conference on Machine Learning Applications, 2017. (Best Paper Award!)
- Bootstrapped Adaptive Threshold Selection for Statistical Model Selection and Estimation. Bouchard, K.E.; arXiv.stat.ML; April, 2015.
Standardization of data formats with Neurodata Without Borders: Neurophysiology
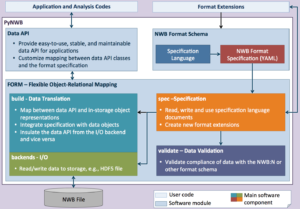
To maximize the return on investment into creation of neuroscience data sets and enhance reproducibility, it is critical to share data through standardized and extensible data model and management solutions. In addition to standardizing data and metadata, support for fast data read/write and high-performance, parallel data analysis are critical to enable labs to keep up with ever growing data volumes. The Neurodata Without Borders: Neurophysiology (NWB-N) effort was an important step towards generating a unified data format for cellular-based neurophysiology data for a multitude of use cases. To enable broad adoption of NWB-N, easily accessible tools and an advanced software strategy aimed at facilitating the use, extension, integration, and maintenance of NWB-N are critically needed. With PyNWB we have developed a new modular software architecture and API to enable users/developers to efficiently interact with the NWB data format, format files, and specifications. Importantly this architecture decouples the various aspects of NWB:N: 1) the specification language, 2) format specification, 3) storage backend, and 4) data API. This allows each to be used and maintained independently. Through its Flexible Object-Relational Mapping functionality, FORM allows us to decouple the data API, format specification, and I/O backends from each other enabling the flexible design of: 1) advanced user APIs, 2) new I/O backends, and 3) data formats and extensions. The FORM module defines a general library for creating scientific data formats and builds the core infrastructure for PyNWB.
Related Publications:
- The Neurodata Without Borders ecosystem for neurophysiological data science. Rubel, O., ….., Bouchard, K.E. eLife, 2022
- HDMF: Hierarchical Data Modeling Framework for Modern Science Data Standards. Tritt, A., Rübel, O., Dichter, B., Ly, R., Chang, E., Kang, D., Frank, L., Bouchard, K.E.; IEEE Big Data, 2019.
- NWB:N 2.0: An Accessible Data Standard for Neurophysiology. Rübel,O., Tritt., A.J., …., Bouchard, K.E.https://www.biorxiv.org/content/10.1101/523035v1
- Methods for Specifying Scientific Data Standards and Modeling Relationships with Applications to Neuroscience. Rübel, O., Dougherty, M., Prabhat, Denes, P., Conant, D., Chang, E.F., Bouchard, K.E.; Front Neuroinform. Nov., 2016;
- High-Performance Computing in Neuroscience for Data-Driven Discovery, Integration, and Dissemination. Bouchard, K.E.*, et al., Neuron, Oct. 2016
Map of vocal tract articulators in human sensorimotor cortex during speech production
 Human speech sounds (e.g. ‘shee’; far left plot) are generated by the rapid, coordinated movement of the vocal tract articulators (i.e. Lips, Jaw, Tongue, and Larynx; center left plot). We used high-density electrocorticography (ECoG) arrays to record neural activity directly from the surface of speech sensorimotor cortex (center right plot) while neurosurgical patients spoke consonant-vowel syllables. Analysis of neural activity revealed that speech articulators are represented in distinct spatial locations (far right plot). The relative location of the different representations were ordered as Larynx, Tongue, Jaw, Lips, Larynx. This was the first time all vocal tract articulators were simultaneously mapped on the human brain during the act of speaking.
Human speech sounds (e.g. ‘shee’; far left plot) are generated by the rapid, coordinated movement of the vocal tract articulators (i.e. Lips, Jaw, Tongue, and Larynx; center left plot). We used high-density electrocorticography (ECoG) arrays to record neural activity directly from the surface of speech sensorimotor cortex (center right plot) while neurosurgical patients spoke consonant-vowel syllables. Analysis of neural activity revealed that speech articulators are represented in distinct spatial locations (far right plot). The relative location of the different representations were ordered as Larynx, Tongue, Jaw, Lips, Larynx. This was the first time all vocal tract articulators were simultaneously mapped on the human brain during the act of speaking.
Related Publications:
- Functional organization of human sensorimotor cortex for speech articulation. Bouchard, K.E., Mesgarani, N., Johnson, K., Chang, E.F.; Nature Article, Feb., 2013.
Context dependent cortical network dynamics for syllable production
 Speech is the quintessential sequential behavior. The cortical dynamics generating these sequences is largely unknown. We used dimensionality reduction (e.g. PCA, LDA) to map the high-dimensional network activity (ECoG data) to a lower dimensional space (‘cortical state-space’) that faithfully retains the structure of the entire network. We first examined state-space dynamics giving rise to different sets of consonant-vowel syllables (left plot). This revealed periodic dynamics during the network transition between distinct states for consonants and vowels. Consonants and vowels were further sub-specified for consonant and vowel features. The organization of phonetic features in the state-space emphasized the articulatory differences between phonemes, but contained additional structure. We then examined the detailed network dynamics with which single phonemes (e.g. /g/ and /u/) transitioned to/from multiple other phonemes on a single-trial basis (right plot). This revealed that the state-space trajectories for single phonemes are biased towards the state-space locations of adjacent phonemes. This bias effectively minimizes the distance the network travels between adjacent states, in agreement with predictions of optimal control theory.
Speech is the quintessential sequential behavior. The cortical dynamics generating these sequences is largely unknown. We used dimensionality reduction (e.g. PCA, LDA) to map the high-dimensional network activity (ECoG data) to a lower dimensional space (‘cortical state-space’) that faithfully retains the structure of the entire network. We first examined state-space dynamics giving rise to different sets of consonant-vowel syllables (left plot). This revealed periodic dynamics during the network transition between distinct states for consonants and vowels. Consonants and vowels were further sub-specified for consonant and vowel features. The organization of phonetic features in the state-space emphasized the articulatory differences between phonemes, but contained additional structure. We then examined the detailed network dynamics with which single phonemes (e.g. /g/ and /u/) transitioned to/from multiple other phonemes on a single-trial basis (right plot). This revealed that the state-space trajectories for single phonemes are biased towards the state-space locations of adjacent phonemes. This bias effectively minimizes the distance the network travels between adjacent states, in agreement with predictions of optimal control theory.
Related Publications:
- Functional organization of human sensorimotor cortex for speech articulation. Bouchard, K.E., Mesgarani, N., Johnson, K., Chang, E.F.; Nature Article, Feb., 2013.
- Cortical control of vowel formants and co-articulation by human sensorimotor cortex. Bouchard, K.E., Chang, E.F.; J. Neuroscience, Sept., 2014.
Towards brain-machine interfaces: single-trial decoding of produced speech
 Brain-machine interfaces (BMI) involve deriving a mathematical mapping from neural signals (brain) to the control of an external effector (machine). There are a large variety of clinical and basic science applications BMIs. However, current performance is relatively modest, especially for speech. Our studies are currently the state-of-the-art in single-trial decoding of speech from human sensorimotor cortex. Together, other results suggest a novel, mixed continuous/discrete approach to a speech prosthetic.
Brain-machine interfaces (BMI) involve deriving a mathematical mapping from neural signals (brain) to the control of an external effector (machine). There are a large variety of clinical and basic science applications BMIs. However, current performance is relatively modest, especially for speech. Our studies are currently the state-of-the-art in single-trial decoding of speech from human sensorimotor cortex. Together, other results suggest a novel, mixed continuous/discrete approach to a speech prosthetic.
Continuous Decoding of Vowel Acoustics. The speech sensorimotor cortex controls the kinematics of the vocal tract articulators. In general, the relationship between vocal tract kinematics and produced acoustics is a many-to-one mapping, and is thus a mathematically degenerate problem. However, the acoustics of vowels during the steady state is more directly related to acoustics than for most other sounds. Therefore, we utilized a novel approach to statistical regularization to decode the produced continuous acoustics of three cardinal vowels (/a/ (‘aa’), /i/ ‘ee’, /u/ ‘oo’) from the concurrently recorded neural activity (left plot). We were able to predict the acoustics of produced vowels on a single-trial basis with extremely high accuracy. This is currently the state-of-the-art in published continuous speech decoding.
Continuous Decoding of Articulator Kinematics. Although we have demonstrated the state-of-the-art in continuous decoding of vowel acoustics, our ability to decode the acoustics of other speech sounds, such as consonants is relatively poor. As discussed above, this in part reflects the fact that the sensorimotor cortex controls the vocal tract articulators, and the mapping from articulators to acoustics is, in general, degenerate. However, simultaneous measurement of all vocal tract articulators is challenging, especially in the clinical setting in which our ECoG recordings are taken. To overcome this challenge, we pioneered a novel, multi-modal system for real-time tracking of all vocal tract articulators during speech production compatible with ECoG recordings in the hospital. Preliminary analysis of lip kinematics has demonstrated the capacity to predict the lip aperature with high fidelity (center). This is the first time that vocal tract articulator kinematics have been directly decoded from human brain recordings.
Deep Neural Nets for Syllable Classification: In contrast to the continuous decoding described above, the most common approach to speech prostheses in the literature treats speech as a sequence of categorical tokens, and attempts to classify these tokens from the recorded neural activity. Most recently, we have been using deep neural networks to classify speech syllables (right). Our initial results are far surpassing the current state-of-the-art published results for speech classification. These results suggest that deep neural networks will be fruitful avenues for neural prosthetics.
Related Publications:
- Neural decoding of spoken vowels from human sensory-motor cortex with high-density electrocorticography. Bouchard, K.E., Chang, E.F.; IEEE, EMBC, Aug., 2014.
- Cortical control of vowel formants and co-articulation by human sensorimotor cortex. Bouchard, K.E., Chang, E.F.; J. Neuroscience, Sept., 2014.
- High-resolution, non-invasive imaging of upper vocal tract articulators compatible with human brain recordings. Bouchard, K.E.*, Conant, D.*, Anumanchipalli, G., Dichter, B.; Johnson, K., Chang, E.F.; PLoS One. 2016
- Decoding speech from human ECoG with deep networks. Livezey, J.*, Anumanchipalli, G.K.*, Prabhat, Bouchard, K.E.** Chang, E.F.**; NIPS Poster; June, 2015. **: co-senior authors
- Deep-learning as a data analysis tool for systems neuroscience. Livezey, J.*, Bouchard, K.E.*$, Chang, E.F.$; PLoS Computational Biology, 15(9): e1007091. Sept, 2019. *: co-first authors; $: co-senior authors
Latent structure of spatio-temporal patterns of neural activity revealed by DNNs
Brain computations are non-linear functions operating on spatio-temporal patterns of neural activity. However, most methods used to understand brain computation are linear, inherently limiting the capacity to extract structure from neural recordings. This is an issue not only for optimization of BMIs, but also for understanding brain computations. As DNNs are essentially adaptive bases function, non-linear function approximators, it is possible that they can extract structure from noisy, single-trial neural recordings that reveal important organization of representations. We examined the structure of network output to more fully understand the organization of syllable representations in vSMC.To the left, we show the average confusion matrix resulting from the output of the softmax layer of the fully connected network (i.e. before binary classification), with target syllables arranged along rows and predicted syllable across columns. The syllables are ordered according to the results of agglomerative hierarchical clustering using Ward’s method. To the right is a bar-plot of the mean accuracy with which a specific syllable was correctly classified. Note that the syllable with worst accuracy is the one with the smallest number of examples in the dataset. At the highest level, syllables seem to be confused only within the articulator involved (lips, back tongue, or front tongue) in the syllable. This is followed by a characterization of the place of articulation within each articulator (bilabial, labio-dental, etc.). At the lowest level there seems to be a clustering across the vowel categories that capture the general shape of the vocal tract in producing the syllable. These results demonstrate the capacity of deep networks to reveal important structure in single-trial neural recordings that is not recoverable with other methods.
Related Publications:
- Decoding speech from human ECoG with deep networks. Livezey, J.*, Anumanchipalli, G.K., Prabhat, Bouchard, K.E.** Chang, E.F.**; NIPS Poster; June, 2015. **: co-senior authors
- Deep-learning as a data analysis tool for systems neuroscience. Livezey, J.*, Bouchard, K.E.*$, Chang, E.F.$; PLoS Computational Biology, 15(9): e1007091. Sept, 2019. *: co-first authors; $: co-senior authors
Sparse sensorimotor representations of articulators
In contrast to our understanding of motor systems, our understanding of sensory systems is relatively advanced. The concept of sparsity has proven central to a theoretical understanding of sensory processing. A population representation is said to be sparse if only a small number of elements are active at any moment in time. Mathematically, sparse representations are often statistically independent. We applied unsupervised ICA to the spatio-temporal patterns of neural activity associated with the different syllables in our data set, and simply visualized the resulting components. We observed two distinct types of sparse representations in sensorimotor cortex activity. Several ICs that accounted for the largest amount of variance were activated by all syllables that engaged the same articulator (left). Specifically, we found that IC2 was activated during syllables with labial consonants (green), IC4 was activated during syllables with dorsal tongue consonants, and IC6 was activated during syllables with coronal tongue consonants. This suggests a sparse representation in an articulator basis. Together, these data support the notion of sparse sensorimotor representations during behavior, and are the first observations of their type mammalian motor cortex.
Related Publications:
- Sparse coding of ECoG signals identifies interpretable components for speech control in human sensorimotor cortex. Bouchard, K.E.*, Bujan, A.F., Chang, E.F., Sommer, F.T.; IEEE, EMBC, Aug., 2017.
Encoding and integration of learned probabilistic birdsong sequences
M any complex behaviors, such as human speech and birdsong, reflect a set of categorical actions that can be flexibly organized into variable sequences. However, little is known about how the brain encodes the probabilities of such sequences. Behavioral sequences are typically characterized by the probability of transitioning from a given action to any subsequent action (forward probability; left plot). In contrast, we hypothesized that neural circuits might encode the probability of transitioning to a given action from any preceding action (backward probability; left plot). To test whether backward probability is encoded in the nervous system, we investigated how auditory-motor neurons in vocal premotor nucleus HVC of songbirds encode different probabilistic characterizations of produced syllable sequences. We recorded responses to auditory playback of pseudo-randomly sequenced syllables from the bird’s repertoire, and found that variations in responses to a given syllable could be explained by a positive linear dependence on the backward probability of preceding sequences (center plot). Furthermore, backward probability accounted for more response variation than other probabilistic characterizations, including forward probability (center plot). Finally, we found that responses integrated over more than 7-10 syllables (~700-1000 ms) with the sign, gain and temporal extent of integration depending on backward probability (right plot). Our results demonstrate that backward probability is encoded in sensory-motor circuitry of the song-system, and suggest that encoding of backward probability is a general feature of sensory-motor circuits.
any complex behaviors, such as human speech and birdsong, reflect a set of categorical actions that can be flexibly organized into variable sequences. However, little is known about how the brain encodes the probabilities of such sequences. Behavioral sequences are typically characterized by the probability of transitioning from a given action to any subsequent action (forward probability; left plot). In contrast, we hypothesized that neural circuits might encode the probability of transitioning to a given action from any preceding action (backward probability; left plot). To test whether backward probability is encoded in the nervous system, we investigated how auditory-motor neurons in vocal premotor nucleus HVC of songbirds encode different probabilistic characterizations of produced syllable sequences. We recorded responses to auditory playback of pseudo-randomly sequenced syllables from the bird’s repertoire, and found that variations in responses to a given syllable could be explained by a positive linear dependence on the backward probability of preceding sequences (center plot). Furthermore, backward probability accounted for more response variation than other probabilistic characterizations, including forward probability (center plot). Finally, we found that responses integrated over more than 7-10 syllables (~700-1000 ms) with the sign, gain and temporal extent of integration depending on backward probability (right plot). Our results demonstrate that backward probability is encoded in sensory-motor circuitry of the song-system, and suggest that encoding of backward probability is a general feature of sensory-motor circuits.
Related Publications:
- Neural encoding and integration of learned probabilistic sequences in avian sensory-motor circuitry. Bouchard, K.E., Brainard, M.S.; J. Neuroscience, Nov., 2013.
Predictive neural dynamics for learned temporal and sequential statistics
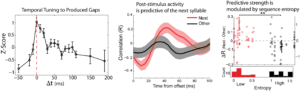 Predicting future events is a critical computation for both perception and behavior. Despite the essential nature of this computation, there are few studies demonstrating neural activity that predicts specific events in learned, probabilistic sequences. Here, we test the hypotheses that the dynamics of internally generated neural activity are predictive of future events and are structured by the learned temporal-sequential statistics of those events. We recorded neural activity in Bengalese finch sensory-motor area HVC in response to playback of sequences from individuals’ songs, and examined the neural activity that continued after stimulus offset. We found that auditory responses to playback of syllable sequences are tuned to the produced timing between syllables. Furthermore, post-stimulus neural activity induced by sequence playback resembles neural responses to the next syllable in the sequence when that syllable is predictable, but not when the next syllable is uncertain. Our results demonstrate that the dynamics of internally generated HVC neural activity are predictive of the learned temporal-sequential structure of produced song and that the strength of this prediction is modulated by uncertainty.
Predicting future events is a critical computation for both perception and behavior. Despite the essential nature of this computation, there are few studies demonstrating neural activity that predicts specific events in learned, probabilistic sequences. Here, we test the hypotheses that the dynamics of internally generated neural activity are predictive of future events and are structured by the learned temporal-sequential statistics of those events. We recorded neural activity in Bengalese finch sensory-motor area HVC in response to playback of sequences from individuals’ songs, and examined the neural activity that continued after stimulus offset. We found that auditory responses to playback of syllable sequences are tuned to the produced timing between syllables. Furthermore, post-stimulus neural activity induced by sequence playback resembles neural responses to the next syllable in the sequence when that syllable is predictable, but not when the next syllable is uncertain. Our results demonstrate that the dynamics of internally generated HVC neural activity are predictive of the learned temporal-sequential structure of produced song and that the strength of this prediction is modulated by uncertainty.
Related Publications:
- Auditory induced neural dynamics in sensory-motor circuitry predict learned temporal and sequential statistics of birdsong. Bouchard, K.E., Brainard, M.S.; Proceedings of the National Academy of Science, Aug., 2016.
Encoding of Learned Probabilistic Speech Sequences in Humans
 Sensory processing involves identification of stimulus features, but also integration with the surrounding sensory and cognitive context. Previous work in animals and humans has shown fine-scale sensitivity to context in the form of learned knowledge about the statistics of the sensory environment, including relative probabilities of discrete units in a stream of sequential auditory input. These statistics are a defining characteristic of one of the most important sequential signals humans encounter: speech. For speech, extensive exposure to a language tunes listeners to the statistics of sound sequences. To address how speech sequence statistics are neurally encoded, we used high-resolution direct cortical recordings from human superior temporal gyrus (right plot) as subjects listened to words and nonwords with varying transition probabilities between sound segments (center plot). In addition to their sensitivity to acoustic features (including contextual features like coarticulation), we found that neural responses dynamically encoded the language-level probability of both preceding and upcoming speech sounds. Transition probability first negatively modulated neural responses, followed by positive modulation of neural responses, consistent with coordinated predictive and retrospective recognition processes, respectively (right plot). These results demonstrate that sensory processing of deeply learned stimuli involves integrating physical stimulus features with their contextual sequential structure.
Sensory processing involves identification of stimulus features, but also integration with the surrounding sensory and cognitive context. Previous work in animals and humans has shown fine-scale sensitivity to context in the form of learned knowledge about the statistics of the sensory environment, including relative probabilities of discrete units in a stream of sequential auditory input. These statistics are a defining characteristic of one of the most important sequential signals humans encounter: speech. For speech, extensive exposure to a language tunes listeners to the statistics of sound sequences. To address how speech sequence statistics are neurally encoded, we used high-resolution direct cortical recordings from human superior temporal gyrus (right plot) as subjects listened to words and nonwords with varying transition probabilities between sound segments (center plot). In addition to their sensitivity to acoustic features (including contextual features like coarticulation), we found that neural responses dynamically encoded the language-level probability of both preceding and upcoming speech sounds. Transition probability first negatively modulated neural responses, followed by positive modulation of neural responses, consistent with coordinated predictive and retrospective recognition processes, respectively (right plot). These results demonstrate that sensory processing of deeply learned stimuli involves integrating physical stimulus features with their contextual sequential structure.
Related Publications:
- Dynamic encoding of speech sequence probability in human temporal cortex. Leonard, M.K., Bouchard, K.E., Tang, C; Chang, E.F.; J. Neuroscience, May, 2015.
Hebbian mechanisms of probabilistic sequence encoding in neural networks
 The majority of distinct motor (and sensory) events occur as temporally ordered sequences with rich probabilistic structure. Sequences can be characterized by the probability of transitioning from the current state to upcoming states (forward probability), as well as the probability of having transitioned to the current state from previous states (backward probability). Despite the prevalence of probabilistic sequencing of both sensory and motor events, the Hebbian mechanisms (left plot) that mold synapses to reflect the statistics of experienced probabilistic sequences are not well understood. We demonstrated that to stably reflect the conditional probability of a neuron’s inputs and outputs, local Hebbian plasticity requires a balance of the magnitude of competitive forces and homogenizing forces (center plot). We showed through analytic calculations and numerical simulations that asymmetric Hebbian plasticity (correlation, covariance and STDP) with pre-synaptic competition can develop synaptic weights equal to the conditional forward transition probabilities present in the input sequence (right plot). In contrast, post-synaptic competition can develop synaptic weights proportional to the conditional backward probabilities of the same input sequence (right plot). Together, these results demonstrate a simple correspondence between the biophysical organization of neurons, the site of synaptic competition, and the temporal flow of information encoded in synaptic weights by Hebbian plasticity while highlighting the necessity of balancing learning forces to accurately encode probability distributions.
The majority of distinct motor (and sensory) events occur as temporally ordered sequences with rich probabilistic structure. Sequences can be characterized by the probability of transitioning from the current state to upcoming states (forward probability), as well as the probability of having transitioned to the current state from previous states (backward probability). Despite the prevalence of probabilistic sequencing of both sensory and motor events, the Hebbian mechanisms (left plot) that mold synapses to reflect the statistics of experienced probabilistic sequences are not well understood. We demonstrated that to stably reflect the conditional probability of a neuron’s inputs and outputs, local Hebbian plasticity requires a balance of the magnitude of competitive forces and homogenizing forces (center plot). We showed through analytic calculations and numerical simulations that asymmetric Hebbian plasticity (correlation, covariance and STDP) with pre-synaptic competition can develop synaptic weights equal to the conditional forward transition probabilities present in the input sequence (right plot). In contrast, post-synaptic competition can develop synaptic weights proportional to the conditional backward probabilities of the same input sequence (right plot). Together, these results demonstrate a simple correspondence between the biophysical organization of neurons, the site of synaptic competition, and the temporal flow of information encoded in synaptic weights by Hebbian plasticity while highlighting the necessity of balancing learning forces to accurately encode probability distributions.
Related Publications:
- Role of the site of synaptic competition and the balance of learning forces for Hebbian encoding of probabilistic Markov sequences. Bouchard, K.E., Ganguly, S., Brainard, M.S.; Front. in Comp. Neuro., July, 2015.


Recent Comments